
| We developed methods to insert single copy transgenes at random sites (miniMos), to insert single copy transgenes at known sites permissive for gene expression (MosSCI), and to modify the genome at any site using a high-throughput CRISPR methodology in C. elegans (SapTrap). | |
|
|
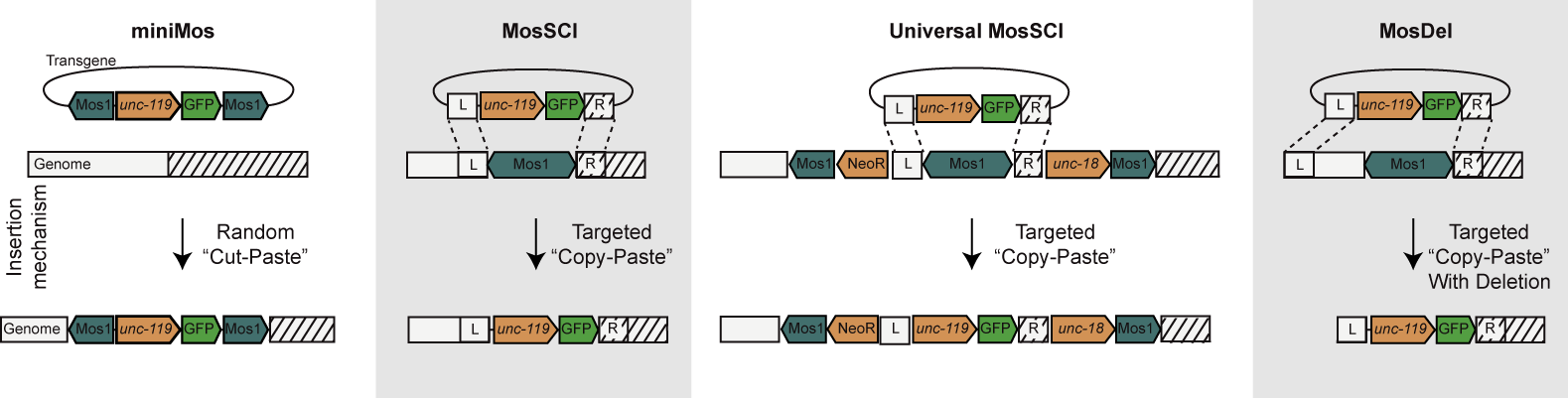
| Random transgene insertions (miniMos). Previously we developed methods to mobilize the Drosophila Mos1 transposon in the C. elegans genome. However, Mos1 transposons carrying heterologous DNA could not be mobilized. We designed a recombinant Mos1 transposon that can hop large pieces of heterologous DNA into the genome. We are using miniMos with the Fire lab to randomly probe permissive versus silent regions of the genome. Targeted single copy insertions (MosSCI). Using standard methods, transgenes in C. elegans are composed of large, repetitive extrachromosomal arrays that are overexpressed or improperly expressed. In the last five years, we have developed efficient methods to insert single-copy transgenes into multiple known sites in the genome that are permissive for expression of transgenes. Even with the advent of CRISPR, MosSCI remains the preferred method for single copy transgenes; these reagents have been requested 2,872 times from Addgene. Targeted single copy insertions at multiple sites (UniversalMosSCI). In Drosophila, the PhiC31 system is used to efficiently insert transgenes into pre-defined sites in the genome. This system does not work in C. elegans. We have combined the miniMos and MosSCI systems to engineer an effective functionally equivalent system. We use a specially designed miniMos vector that contains within it a wild-type Mos transposon flanked by short universal homology arms. We have generated many worm strains each with a unique insertion of this transposon into ints genome. These strains can be used with standard MosSCI vectors to insert a transgene into any of these multiple strains to insert the same transgene into a variety of genomic contexts. Targeted genomic deletions (MosDel). We have shown that by directing one of the homology arms to a distant site in the genome the MosSCI system can be used to generate precisely engineered deletions of the genome adjacent to a Mos1 insertion site. This system was developed prior to CRISPR/Cas9 based genome taregeting systems and was used to generate a number of informative gene deletions. | |
 |
|
|---|---|
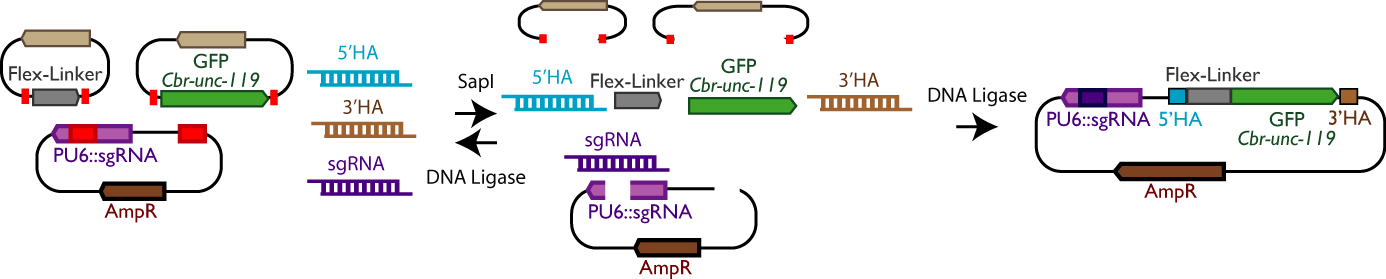
| High-throughput CRISPR (SapTrap). CRISPR/Cas9 can be used to alter or tag genes at any locus. However, DNA reagent construction and identification of modified strains are laborious. We developed a high-throughput method for CRISPR which features a simple and novel selection scheme and greatly simplifies the design and construction of DNA reagents for individual edits. |
>
Frøkjær-Jensen, C., M.W. Davis, M. Sarov, J. Taylor, S. Flibotte, M. LaBella, A. Pozniakovsky, D.G Moerman, E.M. Jorgensen. 2014. Random and targeted integration of transgenes in C. elegans using a modified Mos1 transposon. Nature Methods (in press). |
|
Frøkjær-Jensen, C., M.W. Davis, M. Ailion, E.M. Jorgensen. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nature Methods 9, p117-118. |
|
Zeiser E, Frøkjær-Jensen C, Jorgensen E, Ahringer J. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One. 2011;6(5):e20082. |
|
Frøkjær-Jensen, C., M.W. Davis, G. Hollopeter, J. Taylor, T. Harris, P. Nix, R. Lofgren, M. Bastiani, D.G. Moerman, E.M. Jorgensen. 2010. Targeted gene deletions in C. elegans. Nature Methods 7:451-453. |
|
Frøkjær-Jensen, C., M.W. Davis, C.E. Hopkins, B. Newmann, J.M. Thummel, S-P. Olesen, M. Grunnet and E.M. Jorgensen. 2008. Single copy insertion of transgenes in C. elegans. Nature Genetics 40, p1375-1383. |
|
Robert, V.J., M.W. Davis, E.M. Jorgensen and J-L Bessereau. 2008. Gene conversion and end-joining repair double-strand breaks in the C. elegans germline. Genetics 180, p673 - 679. |
|
Davis, M.W., J. Morton, D. Carroll and E.M. Jorgensen. 2008. Gene activation using FLP recombinase in C. elegans. PLoS Genetics 4(3) e1000028. |
|
Morton, J., M.W. Davis, E.M. Jorgensen, and D. Carroll. 2006. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in C. elegans somatic cells. Proc Natl Acad Sci USA 103, p16370-16375. |
|
Williams, D.C., T. Boulin, A.F. Ruaud, E.M. Jorgensen, and J.L Bessereau. 2005. Characterization of Mos1-mediated mutagenesis in Caenorhabditis elegans: A method for the rapid identification of mutated genes. Genetics, 169: p1779-1785. |
|
Bessereau, J.L., A. Wright, D.C. Williams, K. Schuske, M.W. Davis and E.M. Jorgensen. 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413, p70-74. |
Web design by Nels Jorgensen
Contact: Ithica2709 at gmail dot com